Harvard: Researchers
at Harvard Medical School and Beth Israel Deaconess Medical Center have
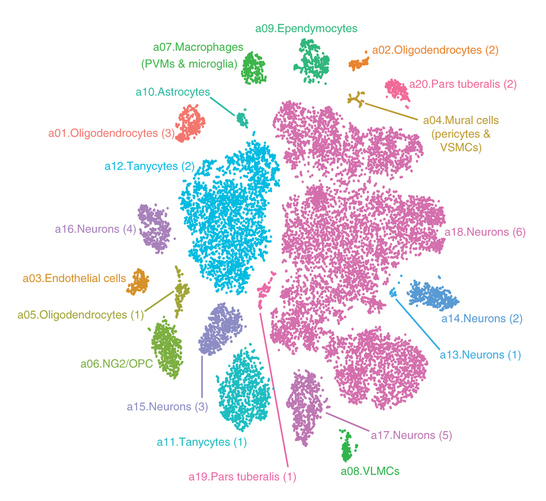
catalogued more than 20,000 brain cells in one region of the mouse
hypothalamus. The study, published in Nature Neuroscience, revealed
some 50 distinct cell types, including a previously undescribed neuron
type that may underlie some of the genetic risk of human obesity. This catalog of cell types marks the first time neuroscientists have
established a comprehensive “parts list” for this area of the brain. The
new information will allow researchers to establish which cells play
what roles.
“A lot of functions have already been mapped to large regions of the brain; for example, we know that the hippocampus is important for memory, and we know the hypothalamus is responsible for basic functions like eating and drinking,” said lead author John N. Campbell, a postdoctoral fellow in the lab of co-corresponding author Bradford Lowell, HMS professor of medicine at Beth Israel Deaconess. “But we don’t know what cell types within those regions are responsible.”
“Now, with the leaps we’ve had in technology,” said Campbell, “we can profile every gene in tens of thousands of individual cells simultaneously and start to test those cell types one by one to figure out their functional roles.”
Each cell in an animal’s body carries the same genetic information. Cells take on specific roles by expressing some genes and silencing others. Drop-Seq technology, developed by study co-authors Steven McCarroll, the Dorothy and Milton Flier Associate Professor of Biomedical Science and Genetics at HMS, and Evan Macosko, HMS assistant professor of psychiatry at Massachusetts General Hospital, makes it possible to assess every gene expressed by individual cells.
The automated process allowed the research team to profile tens of
thousands of cells in the same amount of time it once took to profile
about a dozen cells by hand.
Campbell and colleagues profiled more than 20,000 adult mouse brain cells in the arcuate hypothalamus and the adjoining median eminence, a region of the brain that controls appetite and other vital functions. The cells’ gene expression profiles help scientists determine their functions.
In addition to identifying 50 new cell types, the researchers also profiled the cell types in adult mice under different feeding conditions: eating at will, high-fat diet (energy surplus) and overnight fasting (energy deficit).
The technology allowed the researchers to assess how changes in energy status affected gene expression. The cell types and genes that were sensitive to these changes in energy status provide a number of new targets for obesity treatment.
“Sometimes a cell’s true identity doesn’t come out until you put it through a certain stress,” said co-corresponding author Linus Tsai, HMS assistant professor of medicine at Beth Israel Deaconess. “In fasting conditions, for example, we can see whether there is further diversity within the cell types based on how they respond to important physiologic states.”
Campbell and colleagues have posted their massive data set online, making it available to researchers around the world. The open-source information should accelerate the pace of scientific discovery and shape the questions asked in the field of obesity research.
“The classic way of doing science is to ask questions and test hypotheses,” said Lowell. “But the brain is so complex, we don’t even know how much we don’t know. This information fills in some of the unknowns so we can make new hypotheses. This work will lead to many discoveries that, without these data, people would never have even known to ask the question.”
This work was supported by the National Institutes of Health (grants R01 DK096010, R01 DK089044, R01 DK071051, R01 DK075632, R37 DK053477, BNORC Transgenic Core P30 DK046200, BADERC Transgenic Core P30 DK057521, R01 DK102170, R01 DK085171, R01 DK102173, BNORC Functional Genomics Core P30 DK046200, BADERC Pilot and Feasibility grant NIH 2P30DK057521-16, and F32 DK103387), the Department of Defense (Discovery Award W81XWH-15-1-0251), the American Heart Association (postdoctoral fellowship 14POST20100011), the Lundbeck Foundation and the Benzon Foundation, the Stanley Center for Psychiatric Research and a Stanley-MGH fellowship in psychiatric neuroscience.
Adapted from a Beth Israel Deaconess news release.
“A lot of functions have already been mapped to large regions of the brain; for example, we know that the hippocampus is important for memory, and we know the hypothalamus is responsible for basic functions like eating and drinking,” said lead author John N. Campbell, a postdoctoral fellow in the lab of co-corresponding author Bradford Lowell, HMS professor of medicine at Beth Israel Deaconess. “But we don’t know what cell types within those regions are responsible.”
“Now, with the leaps we’ve had in technology,” said Campbell, “we can profile every gene in tens of thousands of individual cells simultaneously and start to test those cell types one by one to figure out their functional roles.”
Each cell in an animal’s body carries the same genetic information. Cells take on specific roles by expressing some genes and silencing others. Drop-Seq technology, developed by study co-authors Steven McCarroll, the Dorothy and Milton Flier Associate Professor of Biomedical Science and Genetics at HMS, and Evan Macosko, HMS assistant professor of psychiatry at Massachusetts General Hospital, makes it possible to assess every gene expressed by individual cells.
Steve McCarroll and Evan Macosko describe DropSeq in this video.
Campbell and colleagues profiled more than 20,000 adult mouse brain cells in the arcuate hypothalamus and the adjoining median eminence, a region of the brain that controls appetite and other vital functions. The cells’ gene expression profiles help scientists determine their functions.
In addition to identifying 50 new cell types, the researchers also profiled the cell types in adult mice under different feeding conditions: eating at will, high-fat diet (energy surplus) and overnight fasting (energy deficit).
The technology allowed the researchers to assess how changes in energy status affected gene expression. The cell types and genes that were sensitive to these changes in energy status provide a number of new targets for obesity treatment.
“Sometimes a cell’s true identity doesn’t come out until you put it through a certain stress,” said co-corresponding author Linus Tsai, HMS assistant professor of medicine at Beth Israel Deaconess. “In fasting conditions, for example, we can see whether there is further diversity within the cell types based on how they respond to important physiologic states.”
“The brain is so complex, we don’t even know how much we don’t know. This information fills in some of the unknowns so we can make new hypotheses.” —Bradford LowellFinally, the scientists analyzed previous human genome-wide association studies (GWAS) that revealed gene variants linked to obesity. Noting which brain cell types express such obesity-related genes, the researchers implicated two novel neuron types in the genetic control of body weight.
Campbell and colleagues have posted their massive data set online, making it available to researchers around the world. The open-source information should accelerate the pace of scientific discovery and shape the questions asked in the field of obesity research.
“The classic way of doing science is to ask questions and test hypotheses,” said Lowell. “But the brain is so complex, we don’t even know how much we don’t know. This information fills in some of the unknowns so we can make new hypotheses. This work will lead to many discoveries that, without these data, people would never have even known to ask the question.”
This work was supported by the National Institutes of Health (grants R01 DK096010, R01 DK089044, R01 DK071051, R01 DK075632, R37 DK053477, BNORC Transgenic Core P30 DK046200, BADERC Transgenic Core P30 DK057521, R01 DK102170, R01 DK085171, R01 DK102173, BNORC Functional Genomics Core P30 DK046200, BADERC Pilot and Feasibility grant NIH 2P30DK057521-16, and F32 DK103387), the Department of Defense (Discovery Award W81XWH-15-1-0251), the American Heart Association (postdoctoral fellowship 14POST20100011), the Lundbeck Foundation and the Benzon Foundation, the Stanley Center for Psychiatric Research and a Stanley-MGH fellowship in psychiatric neuroscience.
Adapted from a Beth Israel Deaconess news release.