 Purdue: Researchers have introduced a new type of
“super-resolution” microscopy and used it to discover the precise
walking mechanism behind tiny structures made of DNA that could find
biomedical and industrial applications. The researchers also demonstrated how the “DNA
walker” is able to release an anticancer drug, representing a potential
new biomedical technology, said Jong Hyun Choi, an associate professor of mechanical engineering at Purdue University.
Purdue: Researchers have introduced a new type of
“super-resolution” microscopy and used it to discover the precise
walking mechanism behind tiny structures made of DNA that could find
biomedical and industrial applications. The researchers also demonstrated how the “DNA
walker” is able to release an anticancer drug, representing a potential
new biomedical technology, said Jong Hyun Choi, an associate professor of mechanical engineering at Purdue University.
Synthetic nanomotors and walkers are intricately
designed systems that draw chemical energy from the environment and
convert it into mechanical motion. However, because they are too small
to be observed using conventional light microscopes, researchers have
been unable to learn the precise steps involved in the walking
mechanisms, knowledge essential to perfecting the technology.
“If you cannot resolve or monitor these walkers
in action, you will be unable to understand their mechanical operation,”
Choi said.
He led a Purdue team that has solved this problem
by developing a super-resolution microscopy system designed to study
the DNA walkers. The new findings appeared in the journal Science Advances on Jan. 20.
Researchers around the world are creating
synthetic motors based on DNA and RNA, the genetic materials in cells
that consist of a sequence of four chemical bases: adenine, guanine,
cytosine and thymine. The designs are inspired by natural biological
motors that have evolved to perform specific tasks critical to the
function of cells.
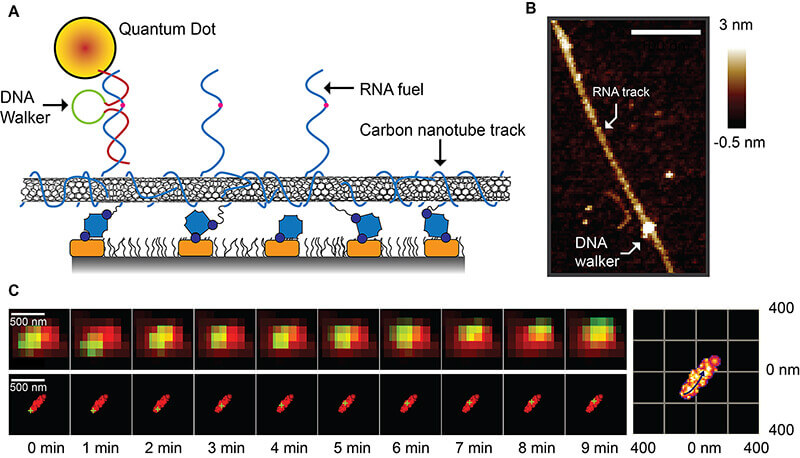
The Purdue researchers have designed a DNA
walking system consisting of an enzymatic core and two arms. The walker
travels along a carbon-nanotube track “decorated” with strands of RNA.
The enzymatic core cleaves off segments of these RNA strands as the
walker continuously moves forward, binding to and harvesting energy from
the RNA. The walker moves in a six-step cycle that repeats as long as
there is RNA fuel.
A new type of “super-resolution” microscopy
has allowed researchers at Purdue University to determine the walking
mechanism behind a DNA walking system that could find biomedical and
industrial applications. The walker (A) travels along a carbon-nanotube
track “decorated” with strands of RNA fuel, which it harvests for
energy. An atomic force microscope image (B) shows the DNA walker
attached to this track. At bottom are raw images taken with the
super-resolution microscope showing the DNA walker (green) traveling
along the track (red). (Purdue University image/Jing Pan)
Download image
A fluorescent nanoparticle is attached
to one arm of the DNA walker, causing it to glow when exposed to light
in the visible part of the spectrum. The carbon-nanotube track also
fluoresces when exposed to light in a portion of the near-infrared
spectrum. Because the new super-resolution microscopy system operates in
both the visible and near-infrared spectra, it is possible to track the
walking mechanism.
The super-resolution technology allows
researchers to resolve structural features far smaller than the
wavelength of visible light, which is normally difficult using
conventional microscopes because of the Abbe diffraction limit,
established by physicist Ernst Abbe in 1873. The limit is about 250
nanometers, which is large compared to the tiny walkers, measuring about
5 nanometers long.
As the DNA walker is exposed to laser light, the
nanoparticle and nanotube flash on and off randomly. These flashes are
captured as numerous fluorescing dots in thousands of imaging frames.
This collection of points is then used to reconstruct the precise motion
of the walker, which moves in a six-step cycle that involves cleaving
portions of the RNA strand and harvesting its energy before moving on to
the next strand.
Findings revealed three primary steps dominate this walking mechanism.
“So, if you can control these three steps within
this walking cycle then you can really study and better control these
walkers,” Choi said. “You can speed them up, you can make them stop and
move in different directions.”
Whereas it previously would have taken 20 hours
or longer to study a complete walking cycle, the new approach speeds the
process to roughly one minute.
The paper was authored by graduate student Jing
Pan; former doctoral student Tae-Gon Cha; graduate students Feiran Li
and Haorong Chen; former undergraduate student Nina A. Bragg; and Choi.
Future research will include work to develop the
drug-release method. The research was funded by the Office of Naval
Research and National Science Foundation.
Writer: Emil Venere, 765-494-4709, venere@purdue.edu
Source: Jong Hyun Choi, 765-496-3562, jchoi@purdue.edu