They’ve identified something that some of those diabetes-linked genetic defects have in common: they seem to change the way certain cells in the pancreas “read” their genes.
The discovery could eventually help lead to more personalized treatments for diabetes. But for now, it’s the first demonstration that many Type 2 diabetes-linked DNA changes have to do with the same DNA-reading molecule. Called Regulatory Factor X, or RFX, it’s a master regulator for a number of genes.
The team reporting the findings in a new paper in the Proceedings of the National Academy of Sciences (link is external) comes from the University of Michigan, National Institutes of Health, Jackson Laboratory for Genomic Medicine, University of North Carolina, and the University of Southern California.
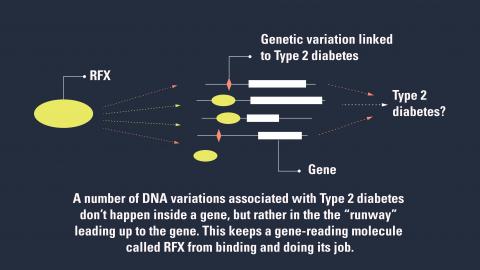
They report that many diabetes-linked DNA changes affect the ability of RFX to bind to specific locations in the genomes of pancreas cell clusters called islets. And that in turn changes the cells’ ability to carry out important functions.
Islets contain the cells that make hormones, including insulin and glucagon, which keep blood sugar balanced in healthy people. In people with diabetes, that regulation goes awry – leading to a range of health problems that can develop over many years.

Stephen C. J. Parker, Ph.D.
Parker is one of four co-senior authors on the paper, which also includes Michael Boehnke, Ph.D., of the U-M School of Public Health’s Department of Biostatistics, Francis Collins, M.D., Ph.D., director of the National Institutes of Health, and Michael L. Stitzel, Ph.D. of the Jackson Laboratory.
Prior to their current faculty positions Parker and Stitzel worked in Collins’ lab at the National Human Genome Research Institute. Parker’s graduate student, Arushi Varshney, is one of the paper’s co-first authors with Laura Scott, Ph.D., and Ryan Welch, Ph.D., of the U-M School of Public Health’s Department of Biostatistics and Michael Erdos, Ph.D., of the National Human Genome Research Institute.
They performed an extensive examination of DNA from islet samples isolated from 112 people. They characterized differences not just in DNA sequences, but also in the way DNA was packaged and modified by epigenetic factors, and the levels of gene expression products that indicated how often the genes had been read and transcribed.
This allowed them to track the “footprints” that RFX and other transcription factors leave on packaged DNA after they have done their job.
RFX and other factors don’t bind directly to the part of a gene that encodes a protein that does a cellular job. Rather, they bind to a stretch of DNA near the gene – a runway of sorts.
But when genetic changes linked to Type 2 diabetes are present, that runway gets disrupted, and RFX can’t bind as it should.
Each DNA change might alter this binding in a different way, leading to a slightly different effect on Type 2 diabetes risk or blood sugar regulation. But the common factor for many of these changes was its effect on the area where RFX is predicted to bind, in the cells of pancreatic islets.
So, says Parker, this shows how the genome – the actual sequence of DNA -- can influence the epigenome, or the factors that influence gene expression.
The researchers note that a deadly form of diabetes seen in a handful of babies born each year may be related to RFX mutations. That condition, called Mitchell-Riley syndrome, involves neonatal diabetes and malformed pancreas, and is known to be caused by a rare autosomal recessive mutation of one form of RFX.
In addition to co-senior and co-first authors listed above, the study’s authors include a range of researchers from several institutions. The study was funded by the National Institutes of Health (HL127984, DK062370, HG000024, DK099240, DK092251, DK093757, DK105561, DK072193, ZIA HG000024). Parker is a 2014 recipient of the American Diabetes Association’s Pathway to Stop Diabetes grant, a type of grant awarded annually by the American Diabetes Association to provide up to $1.625 million to each scientist over a five- to seven-year grant term to spur breakthroughs in clinical science, technology, diabetes care and potential cures. Since launching in 2013 Pathway has awarded more than $36 million to 23 leading scientists.
Reference: PNAS, www.pnas.org/cgi/doi/10.1073/pnas.1621192114 (link is external)